Il modulo networkx dei grafi su Python
Per lavorare con i grafi su Python posso usare il modulo newtorkx. Si tratta di un modulo appositamente pensato per lavorare con i grafi e i digrafi, sia semplici che multipli.
Prima di tutto va installato sul PC usando il comando pip, perché non è incluso nei moduli standard del linguaggio.
pip install networkx
Una volta fatto questo, basta importarlo nel codice e utilizzare tutte le funzioni disponibili all'interno della libreria.
Un esempio di grafo semplice
Per visualizzare graficamente il risultato è necessario importare anche la libreria matplotlib.
import networkx as nx
import matplotlib.pyplot as plt
Questo mi permette di definire un grafico e assegnarlo alla variabile G tramite il metodo Graph().
G = nx.Graph()
Poi aggiungo gli spigoli (o archi) tra i vertici del grafo tramite il metodo add_edge().
G.add_edge('A', 'B')
G.add_edge('B', 'C')
G.add_edge('C', 'A')
G.add_edge('A', 'D')
G.add_edge('B', 'D')
G.add_edge('B', 'E')
G.add_edge('D', 'E')
Ad esempio, add_edge('A','B') aggiunge un collegamento tra il vertice 'A' e il vertice 'B' mentre add_edge('B','C') lo aggiunge tra i vertici 'B' e 'C'.
In questo esempio, non occorre definire i vertici, basta inserire il collegamento e automaticamente vengono inclusi nel grafo.
Tuttavia, se devo inserire un singolo nodo isolato all'interno del grafo, utilizzo il metodo add_node(). Ad esempio G.add_node('F')
G.add_node('F')
Quando il grafo è ultimato, posso stamparlo usando il metodo draw().
nx.draw(G, with_labels=True)
plt.show()
Il risultato è il seguente:
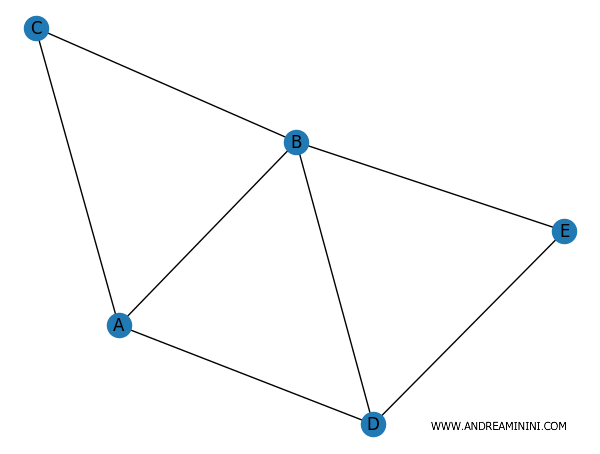
Il modulo networkx non è utile solo perché visualizza graficamente il grafo, questo sarebbe il minimo, ma anche perché mi permette anche di elaborare le principali caratteristiche del grafo.
Ad esempio, posso calcolare il grado di ogni vertice del grafo tramite il metodo degree().
Dove il grado di un vertice è il numero di collegamenti presenti sul vertice stesso. Ad esempio, il vertice A ha 3 spigoli mentre il vertice C ne ha solo 2.
degrees = dict(G.degree())
print(degrees)
{'A': 3, 'B': 4, 'C': 2, 'D': 3, 'E': 2}
Posso anche ottenere le componenti connesse del grafo
components = list(nx.connected_components(G))
print(components)
[{'C', 'D', 'A', 'B', 'E'}]
Trovare il percorso più corto tra due vertici. Ad esempio, tra i vertici A ed E. Uno dei percorsi più corti è A→B→E
shortest_path = nx.shortest_path(G, source='A', target='E')
print(shortest_path)
['A', 'B', 'E']
Posso anche calcolare la centralità dei vertici.
centrality = nx.betweenness_centrality(G)
print(centrality)
{'A': 0.08333333333333333, 'B': 0.3333333333333333, 'C': 0.0, 'D': 0.08333333333333333, 'E': 0.0}
Inoltre, mi permette di calcolare l'albero di copertura minima
T = nx.minimum_spanning_tree(G)
print(T)
Graph with 5 nodes and 4 edges
Un esempio di digrafo
Per creare un grafo orientato (o digrafo) utilizzo il metodo DiGraph().
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
G.add_edge('A', 'B')
G.add_edge('B', 'C')
G.add_edge('C', 'A')
G.add_edge('A', 'D')
G.add_edge('B', 'D')
G.add_edge('B', 'E')
G.add_edge('D', 'E')
nx.draw(G, with_labels=True, arrows=True)
plt.show()
Per inserire i collegamenti si usa lo stesso metodo add_edge() dei grafi semplici (non orientati).
In alternativa, posso usare anche questa sintassi che mi permette di definire più nodi e archi contemporaneamente, usando rispettivamente i metodi add_nodes_from() e add_edges_from()
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
G.add_nodes_from(['A', 'B', 'C', 'D', 'E'])
G.add_edges_from([('A', 'B'), ('B', 'C'), ('C', 'A'), ('A', 'D'), ('B', 'D'), ('B', 'E'), ('D', 'E')])
nx.draw(G, with_labels=True, arrows=True)
plt.show()
In entrambi i casi il grafo viene elaborato dando una direzione agli archi che collegano i nodi del grafo.
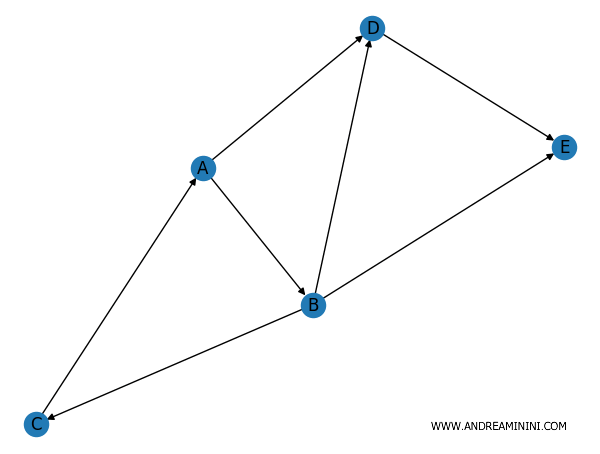
Ora posso calcolare il grado in entrata ovvero il numero di archi che entrano in un nodo.
in_degree = dict(G.in_degree())
print("In-degree:", in_degree)
In-degree: {'A': 1, 'B': 1, 'C': 1, 'D': 2, 'E': 2}
E anche il grado in uscita di ogni nodo.
out_degree = dict(G.out_degree())
print("Out-degree:", out_degre
Out-degree: {'A': 2, 'B': 3, 'C': 1, 'D': 1, 'E': 0}
Posso individuare facilmente le componenti fortemente connesse.
strong_components = list(nx.strongly_connected_components(G))
print(strong_components)
[{'E'}, {'D'}, {'A', 'C', 'B'}]
I grafi multipli (multigrafi)
E' anche possibile definire un grafo multiplo, ossia un grafo in cui i nodi possono avere anche più archi collegati.
Per creare un multigrafo, uso MultiGraph per i grafi non orientati e MultiDiGraph per i grafi orientati.
Ad esempio, definisco un multigrafo non orientato con tre vertici e tre spigoli.
import networkx as nx
MG = nx.MultiGraph()
# Aggiungo i nodi
MG.add_nodes_from([1, 2, 3])
# Aggiungo gli archi multipli
MG.add_edge(1, 2, key='first')
MG.add_edge(1, 2, key='second')
MG.add_edge(2, 3, key='third')
In questo grafo tra i vertici 1 e 2 ci sono due spigoli mentre tra i vertici 2 e 3 c'è un solo spigolo.
Lista dei comandi del modulo
Ecco una lista completa dei metodi, delle funzioni e delle istruzioni del modulo networkx
- AmbiguousSolution
- ArborescenceIterator
- DiGraph
- EdgePartition
- ExceededMaxIterations
- Graph
- GraphMLReader
- GraphMLWriter
- HasACycle
- LCF_graph
- LFR_benchmark_graph
- MultiDiGraph
- MultiGraph
- NetworkXAlgorithmError
- NetworkXError
- NetworkXException
- NetworkXNoCycle
- NetworkXNoPath
- NetworkXNotImplemented
- NetworkXPointlessConcept
- NetworkXTreewidthBoundExceeded
- NetworkXUnbounded
- NetworkXUnfeasible
- NodeNotFound
- NotATree
- PlanarEmbedding
- PowerIterationFailedConvergence
- SpanningTreeIterator
- __builtins__
- __cached__
- __doc__
- __file__
- __loader__
- __name__
- __package__
- __path__
- __spec__
- __version__
- _clear_cache
- _dispatchable
- _lazy_import
- adamic_adar_index
- add_cycle
- add_path
- add_star
- adjacency
- adjacency_data
- adjacency_graph
- adjacency_matrix
- adjacency_spectrum
- adjlist
- algebraic_connectivity
- algebraicconnectivity
- algorithms
- all
- all_neighbors
- all_node_cuts
- all_pairs_all_shortest_paths
- all_pairs_bellman_ford_path
- all_pairs_bellman_ford_path_length
- all_pairs_dijkstra
- all_pairs_dijkstra_path
- all_pairs_dijkstra_path_length
- all_pairs_lowest_common_ancestor
- all_pairs_node_connectivity
- all_pairs_shortest_path
- all_pairs_shortest_path_length
- all_shortest_paths
- all_simple_edge_paths
- all_simple_paths
- all_topological_sorts
- all_triads
- all_triplets
- ancestors
- antichains
- approximate_current_flow_betweenness_centrality
- approximation
- arf_layout
- articulation_points
- assortativity
- astar
- astar_path
- astar_path_length
- asteroidal
- atlas
- attr_matrix
- attr_sparse_matrix
- attracting
- attracting_components
- attribute_assortativity_coefficient
- attribute_mixing_dict
- attribute_mixing_matrix
- attrmatrix
- average_clustering
- average_degree_connectivity
- average_neighbor_degree
- average_node_connectivity
- average_shortest_path_length
- balanced_tree
- barabasi_albert_graph
- barbell_graph
- barycenter
- beamsearch
- bellman_ford_path
- bellman_ford_path_length
- bellman_ford_predecessor_and_distance
- bethe_hessian_matrix
- bethe_hessian_spectrum
- bethehessianmatrix
- betweenness
- betweenness_centrality
- betweenness_centrality_subset
- betweenness_subset
- bfs_beam_edges
- bfs_edges
- bfs_labeled_edges
- bfs_layers
- bfs_layout
- bfs_predecessors
- bfs_successors
- bfs_tree
- biconnected
- biconnected_component_edges
- biconnected_components
- bidirectional_dijkstra
- bidirectional_shortest_path
- binary
- binomial_graph
- binomial_tree
- bipartite
- bipartite_layout
- boundary
- boundary_expansion
- breadth_first_search
- bridges
- broadcasting
- bull_graph
- capacity_scaling
- cartesian_product
- caveman_graph
- cd_index
- center
- centrality
- chain_decomposition
- chains
- check_planarity
- chordal
- chordal_cycle_graph
- chordal_graph_cliques
- chordal_graph_treewidth
- chordless_cycles
- chromatic_polynomial
- chvatal_graph
- circulant_graph
- circular_ladder_graph
- circular_layout
- classes
- classic
- clique
- closeness
- closeness_centrality
- closeness_vitality
- cluster
- clustering
- cn_soundarajan_hopcroft
- cographs
- coloring
- combinatorial_embedding_to_pos
- common_neighbor_centrality
- common_neighbors
- communicability
- communicability_alg
- communicability_betweenness_centrality
- communicability_exp
- community
- complement
- complete_bipartite_graph
- complete_graph
- complete_multipartite_graph
- complete_to_chordal_graph
- components
- compose
- compose_all
- compute_v_structures
- condensation
- conductance
- config
- configuration_model
- connected
- connected_caveman_graph
- connected_components
- connected_double_edge_swap
- connected_watts_strogatz_graph
- connectivity
- constraint
- contracted_edge
- contracted_nodes
- convert
- convert_matrix
- convert_node_labels_to_integers
- core
- core_number
- coreviews
- corona_product
- correlation
- cost_of_flow
- could_be_isomorphic
- covering
- create_empty_copy
- cubical_graph
- current_flow_betweenness
- current_flow_betweenness_centrality
- current_flow_betweenness_centrality_subset
- current_flow_betweenness_subset
- current_flow_closeness
- current_flow_closeness_centrality
- cut_size
- cuts
- cycle_basis
- cycle_graph
- cycles
- cytoscape
- cytoscape_data
- cytoscape_graph
- d_separated
- d_separation
- dag
- dag_longest_path
- dag_longest_path_length
- dag_to_branching
- davis_southern_women_graph
- dedensify
- degree
- degree_alg
- degree_assortativity_coefficient
- degree_centrality
- degree_histogram
- degree_mixing_dict
- degree_mixing_matrix
- degree_pearson_correlation_coefficient
- degree_seq
- degree_sequence_tree
- dense
- dense_gnm_random_graph
- density
- depth_first_search
- desargues_graph
- descendants
- descendants_at_distance
- dfs_edges
- dfs_labeled_edges
- dfs_postorder_nodes
- dfs_predecessors
- dfs_preorder_nodes
- dfs_successors
- dfs_tree
- diameter
- diamond_graph
- difference
- digraph
- dijkstra_path
- dijkstra_path_length
- dijkstra_predecessor_and_distance
- directed
- directed_combinatorial_laplacian_matrix
- directed_configuration_model
- directed_edge_swap
- directed_havel_hakimi_graph
- directed_joint_degree_graph
- directed_laplacian_matrix
- directed_modularity_matrix
- disjoint_union
- disjoint_union_all
- dispersion
- distance_measures
- distance_regular
- dodecahedral_graph
- dominance
- dominance_frontiers
- dominating
- dominating_set
- dorogovtsev_goltsev_mendes_graph
- double_edge_swap
- draw
- draw_circular
- draw_kamada_kawai
- draw_networkx
- draw_networkx_edge_labels
- draw_networkx_edges
- draw_networkx_labels
- draw_networkx_nodes
- draw_planar
- draw_random
- draw_shell
- draw_spectral
- draw_spring
- drawing
- dual_barabasi_albert_graph
- duplication
- duplication_divergence_graph
- eccentricity
- edge_betweenness_centrality
- edge_betweenness_centrality_subset
- edge_bfs
- edge_boundary
- edge_connectivity
- edge_current_flow_betweenness_centrality
- edge_current_flow_betweenness_centrality_subset
- edge_dfs
- edge_disjoint_paths
- edge_expansion
- edge_load_centrality
- edge_subgraph
- edgebfs
- edgedfs
- edgelist
- edges
- effective_graph_resistance
- effective_size
- efficiency
- efficiency_measures
- ego
- ego_graph
- eigenvector
- eigenvector_centrality
- eigenvector_centrality_numpy
- empty_graph
- enumerate_all_cliques
- equitable_color
- equivalence_classes
- erdos_renyi_graph
- estrada_index
- euler
- eulerian_circuit
- eulerian_path
- eulerize
- exception
- expanders
- expected_degree_graph
- extended_barabasi_albert_graph
- fast_could_be_isomorphic
- fast_gnp_random_graph
- faster_could_be_isomorphic
- fiedler_vector
- filters
- find_asteroidal_triple
- find_cliques
- find_cliques_recursive
- find_cycle
- find_induced_nodes
- find_minimal_d_separator
- find_negative_cycle
- florentine_families_graph
- flow
- flow_hierarchy
- flow_matrix
- floyd_warshall
- floyd_warshall_numpy
- floyd_warshall_predecessor_and_distance
- forest_str
- freeze
- from_dict_of_dicts
- from_dict_of_lists
- from_edgelist
- from_graph6_bytes
- from_nested_tuple
- from_numpy_array
- from_pandas_adjacency
- from_pandas_edgelist
- from_prufer_sequence
- from_scipy_sparse_array
- from_sparse6_bytes
- frucht_graph
- fruchterman_reingold_layout
- full_join
- full_rary_tree
- function
- gaussian_random_partition_graph
- general_random_intersection_graph
- generalized_degree
- generate_adjlist
- generate_edgelist
- generate_gexf
- generate_gml
- generate_graphml
- generate_multiline_adjlist
- generate_network_text
- generate_pajek
- generate_random_paths
- generators
- generic
- generic_bfs_edges
- geographical_threshold_graph
- geometric
- geometric_edges
- geometric_soft_configuration_graph
- get_edge_attributes
- get_node_attributes
- gexf
- girth
- global_efficiency
- global_parameters
- global_reaching_centrality
- gml
- gn_graph
- gnc_graph
- gnm_random_graph
- gnp_random_graph
- gnr_graph
- goldberg_radzik
- gomory_hu_tree
- google_matrix
- graph
- graph6
- graph_atlas
- graph_atlas_g
- graph_edit_distance
- graph_hashing
- graphical
- graphmatrix
- graphml
- graphviews
- greedy_color
- grid_2d_graph
- grid_graph
- group
- group_betweenness_centrality
- group_closeness_centrality
- group_degree_centrality
- group_in_degree_centrality
- group_out_degree_centrality
- gutman_index
- harary_graph
- harmonic
- harmonic_centrality
- has_bridges
- has_eulerian_path
- has_path
- havel_hakimi_graph
- heawood_graph
- hexagonal_lattice_graph
- hierarchy
- hits
- hits_alg
- hkn_harary_graph
- hnm_harary_graph
- hoffman_singleton_graph
- house_graph
- house_x_graph
- hybrid
- hypercube_graph
- icosahedral_graph
- identified_nodes
- immediate_dominators
- in_degree_centrality
- incidence_matrix
- incremental_closeness_centrality
- induced_subgraph
- information_centrality
- internet_as_graphs
- intersection
- intersection_all
- intersection_array
- interval_graph
- inverse_line_graph
- is_aperiodic
- is_arborescence
- is_at_free
- is_attracting_component
- is_biconnected
- is_bipartite
- is_branching
- is_chordal
- is_connected
- is_d_separator
- is_digraphical
- is_directed
- is_directed_acyclic_graph
- is_distance_regular
- is_dominating_set
- is_edge_cover
- is_empty
- is_eulerian
- is_forest
- is_frozen
- is_graphical
- is_isolate
- is_isomorphic
- is_k_edge_connected
- is_k_regular
- is_kl_connected
- is_matching
- is_maximal_matching
- is_minimal_d_separator
- is_multigraphical
- is_negatively_weighted
- is_path
- is_perfect_matching
- is_planar
- is_pseudographical
- is_regular
- is_regular_expander
- is_semiconnected
- is_semieulerian
- is_simple_path
- is_strongly_connected
- is_strongly_regular
- is_tournament
- is_tree
- is_triad
- is_valid_degree_sequence_erdos_gallai
- is_valid_degree_sequence_havel_hakimi
- is_valid_directed_joint_degree
- is_valid_joint_degree
- is_weakly_connected
- is_weighted
- isolate
- isolates
- isomorphism
- jaccard_coefficient
- johnson
- join
- join_trees
- joint_degree_graph
- joint_degree_seq
- json_graph
- junction_tree
- k_components
- k_core
- k_corona
- k_crust
- k_edge_augmentation
- k_edge_components
- k_edge_subgraphs
- k_factor
- k_random_intersection_graph
- k_shell
- k_truss
- kamada_kawai_layout
- karate_club_graph
- katz
- katz_centrality
- katz_centrality_numpy
- kemeny_constant
- kl_connected_subgraph
- kneser_graph
- kosaraju_strongly_connected_components
- krackhardt_kite_graph
- ladder_graph
- laplacian
- laplacian_centrality
- laplacian_matrix
- laplacian_spectrum
- laplacianmatrix
- lattice
- lattice_reference
- layout
- lazy_imports
- leda
- les_miserables_graph
- lexicographic_product
- lexicographical_topological_sort
- linalg
- line
- line_graph
- link_analysis
- link_prediction
- load
- load_centrality
- local_bridges
- local_constraint
- local_efficiency
- local_reaching_centrality
- lollipop_graph
- lowest_common_ancestor
- lowest_common_ancestors
- make_clique_bipartite
- make_max_clique_graph
- margulis_gabber_galil_graph
- matching
- max_flow_min_cost
- max_weight_clique
- max_weight_matching
- maximal_independent_set
- maximal_matching
- maximum_branching
- maximum_flow
- maximum_flow_value
- maximum_spanning_arborescence
- maximum_spanning_edges
- maximum_spanning_tree
- maybe_regular_expander
- min_cost_flow
- min_cost_flow_cost
- min_edge_cover
- min_weight_matching
- minimal_d_separator
- minimum_branching
- minimum_cut
- minimum_cut_value
- minimum_cycle_basis
- minimum_edge_cut
- minimum_node_cut
- minimum_spanning_arborescence
- minimum_spanning_edges
- minimum_spanning_tree
- minors
- mis
- mixing
- mixing_dict
- mixing_expansion
- modular_product
- modularity_matrix
- modularity_spectrum
- modularitymatrix
- moebius_kantor_graph
- moral
- moral_graph
- multi_source_dijkstra
- multi_source_dijkstra_path
- multi_source_dijkstra_path_length
- multidigraph
- multigraph
- multiline_adjlist
- multipartite_layout
- mycielski
- mycielski_graph
- mycielskian
- navigable_small_world_graph
- negative_edge_cycle
- neighbor_degree
- neighbors
- network_simplex
- newman_watts_strogatz_graph
- node_attribute_xy
- node_boundary
- node_classification
- node_clique_number
- node_connected_component
- node_connectivity
- node_degree_xy
- node_disjoint_paths
- node_expansion
- node_link
- node_link_data
- node_link_graph
- nodes
- nodes_with_selfloops
- non_edges
- non_neighbors
- non_randomness
- nonisomorphic_trees
- normalized_cut_size
- normalized_laplacian_matrix
- normalized_laplacian_spectrum
- null_graph
- number_attracting_components
- number_connected_components
- number_of_cliques
- number_of_edges
- number_of_isolates
- number_of_nodes
- number_of_nonisomorphic_trees
- number_of_selfloops
- number_of_spanning_trees
- number_of_walks
- number_strongly_connected_components
- number_weakly_connected_components
- numeric_assortativity_coefficient
- nx_agraph
- nx_latex
- nx_pydot
- nx_pylab
- octahedral_graph
- omega
- onion_layers
- operators
- optimal_edit_paths
- optimize_edit_paths
- optimize_graph_edit_distance
- out_degree_centrality
- overall_reciprocity
- pagerank
- pagerank_alg
- pairs
- pajek
- paley_graph
- panther_similarity
- pappus_graph
- parse_adjlist
- parse_edgelist
- parse_gml
- parse_graphml
- parse_leda
- parse_multiline_adjlist
- parse_pajek
- partial_duplication_graph
- partition_spanning_tree
- path_graph
- path_weight
- percolation
- percolation_centrality
- periphery
- petersen_graph
- planar_drawing
- planar_layout
- planarity
- planted_partition_graph
- polynomials
- power
- powerlaw_cluster_graph
- predecessor
- preferential_attachment
- prefix_tree
- prefix_tree_recursive
- product
- projected_graph
- prominent_group
- quotient_graph
- ra_index_soundarajan_hopcroft
- radius
- random_clustered
- random_clustered_graph
- random_cograph
- random_degree_sequence_graph
- random_geometric_graph
- random_graphs
- random_internet_as_graph
- random_k_out_graph
- random_kernel_graph
- random_labeled_rooted_forest
- random_labeled_rooted_tree
- random_labeled_tree
- random_layout
- random_lobster
- random_partition_graph
- random_powerlaw_tree
- random_powerlaw_tree_sequence
- random_reference
- random_regular_expander_graph
- random_regular_graph
- random_shell_graph
- random_spanning_tree
- random_tree
- random_triad
- random_unlabeled_rooted_forest
- random_unlabeled_rooted_tree
- random_unlabeled_tree
- reaching
- read_adjlist
- read_edgelist
- read_gexf
- read_gml
- read_graph6
- read_graphml
- read_leda
- read_multiline_adjlist
- read_pajek
- read_sparse6
- read_weighted_edgelist
- readwrite
- reciprocity
- reconstruct_path
- recursive_simple_cycles
- regular
- relabel
- relabel_gexf_graph
- relabel_nodes
- relaxed_caveman_graph
- reportviews
- rescale_layout
- rescale_layout_dict
- resistance_distance
- resource_allocation_index
- restricted_view
- reverse
- reverse_view
- rich_club_coefficient
- richclub
- ring_of_cliques
- rooted_product
- s_metric
- scale_free_graph
- schultz_index
- second_order
- second_order_centrality
- sedgewick_maze_graph
- selfloop_edges
- semiconnected
- set_edge_attributes
- set_node_attributes
- shell_layout
- shortest_path
- shortest_path_length
- shortest_paths
- shortest_simple_paths
- sigma
- similarity
- simple_cycles
- simple_paths
- simrank_similarity
- single_source_all_shortest_paths
- single_source_bellman_ford
- single_source_bellman_ford_path
- single_source_bellman_ford_path_length
- single_source_dijkstra
- single_source_dijkstra_path
- single_source_dijkstra_path_length
- single_source_shortest_path
- single_source_shortest_path_length
- single_target_shortest_path
- single_target_shortest_path_length
- small
- smallworld
- smetric
- snap_aggregation
- social
- soft_random_geometric_graph
- spanner
- sparse6
- sparsifiers
- spectral_bisection
- spectral_graph_forge
- spectral_layout
- spectral_ordering
- spectrum
- spiral_layout
- spring_layout
- square_clustering
- star_graph
- stochastic
- stochastic_block_model
- stochastic_graph
- stoer_wagner
- strong_product
- strongly_connected
- strongly_connected_components
- strongly_connected_components_recursive
- structuralholes
- subgraph
- subgraph_alg
- subgraph_centrality
- subgraph_centrality_exp
- subgraph_view
- sudoku
- sudoku_graph
- summarization
- swap
- symmetric_difference
- tadpole_graph
- tensor_product
- tetrahedral_graph
- text
- thresholded_random_geometric_graph
- time_dependent
- time_series
- to_dict_of_dicts
- to_dict_of_lists
- to_directed
- to_edgelist
- to_graph6_bytes
- to_latex
- to_latex_raw
- to_nested_tuple
- to_networkx_graph
- to_numpy_array
- to_pandas_adjacency
- to_pandas_edgelist
- to_prufer_sequence
- to_scipy_sparse_array
- to_sparse6_bytes
- to_undirected
- topological_generations
- topological_sort
- total_spanning_tree_weight
- tournament
- transitive_closure
- transitive_closure_dag
- transitive_reduction
- transitivity
- traversal
- tree
- tree_all_pairs_lowest_common_ancestor
- tree_broadcast_center
- tree_broadcast_time
- tree_data
- tree_graph
- trees
- triad_graph
- triad_type
- triadic_census
- triads
- triads_by_type
- triangles
- triangular_lattice_graph
- trivial_graph
- trophic
- trophic_differences
- trophic_incoherence_parameter
- trophic_levels
- truncated_cube_graph
- truncated_tetrahedron_graph
- turan_graph
- tutte_graph
- tutte_polynomial
- unary
- uniform_random_intersection_graph
- union
- union_all
- unweighted
- utils
- vf2pp_all_isomorphisms
- vf2pp_is_isomorphic
- vf2pp_isomorphism
- visibility_graph
- vitality
- volume
- voronoi
- voronoi_cells
- voterank
- voterank_alg
- walks
- watts_strogatz_graph
- waxman_graph
- weakly_connected
- weakly_connected_components
- weighted
- weisfeiler_lehman_graph_hash
- weisfeiler_lehman_subgraph_hashes
- wheel_graph
- wiener
- wiener_index
- windmill_graph
- within_inter_cluster
- write_adjlist
- write_edgelist
- write_gexf
- write_gml
- write_graph6
- write_graphml
- write_graphml_lxml
- write_graphml_xml
- write_latex
- write_multiline_adjlist
- write_network_text
- write_pajek
- write_sparse6
- write_weighted_edgelist
E così via.
